Introduction
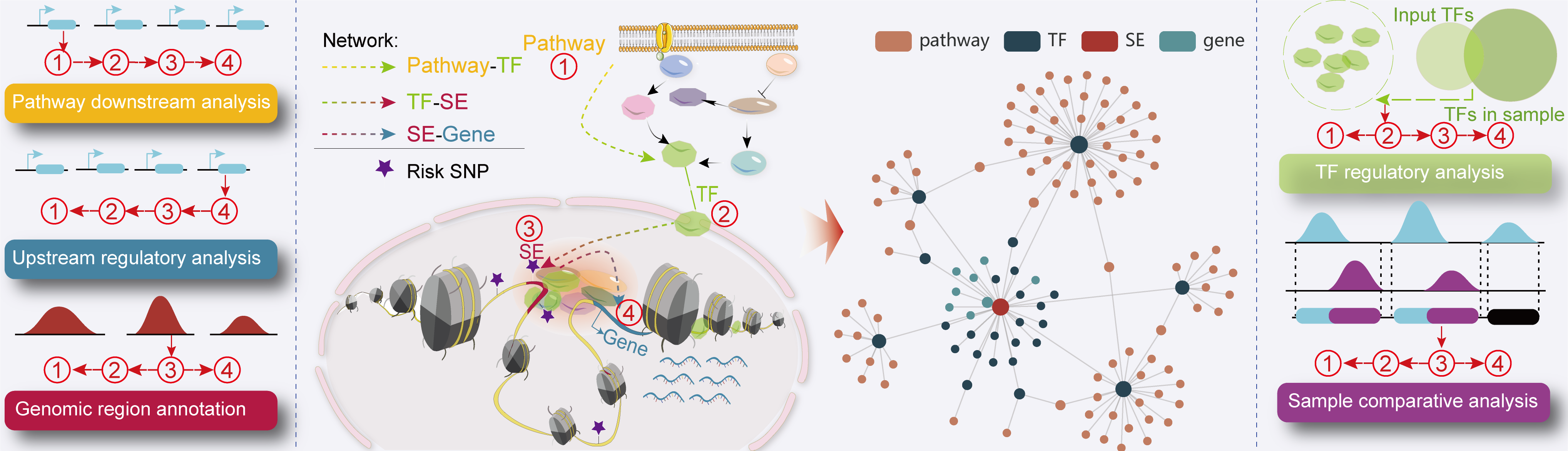
We designed a novel web server, SEanalysis 2.0, which provides a comprehensive super-enhancers regulatory network analysis,
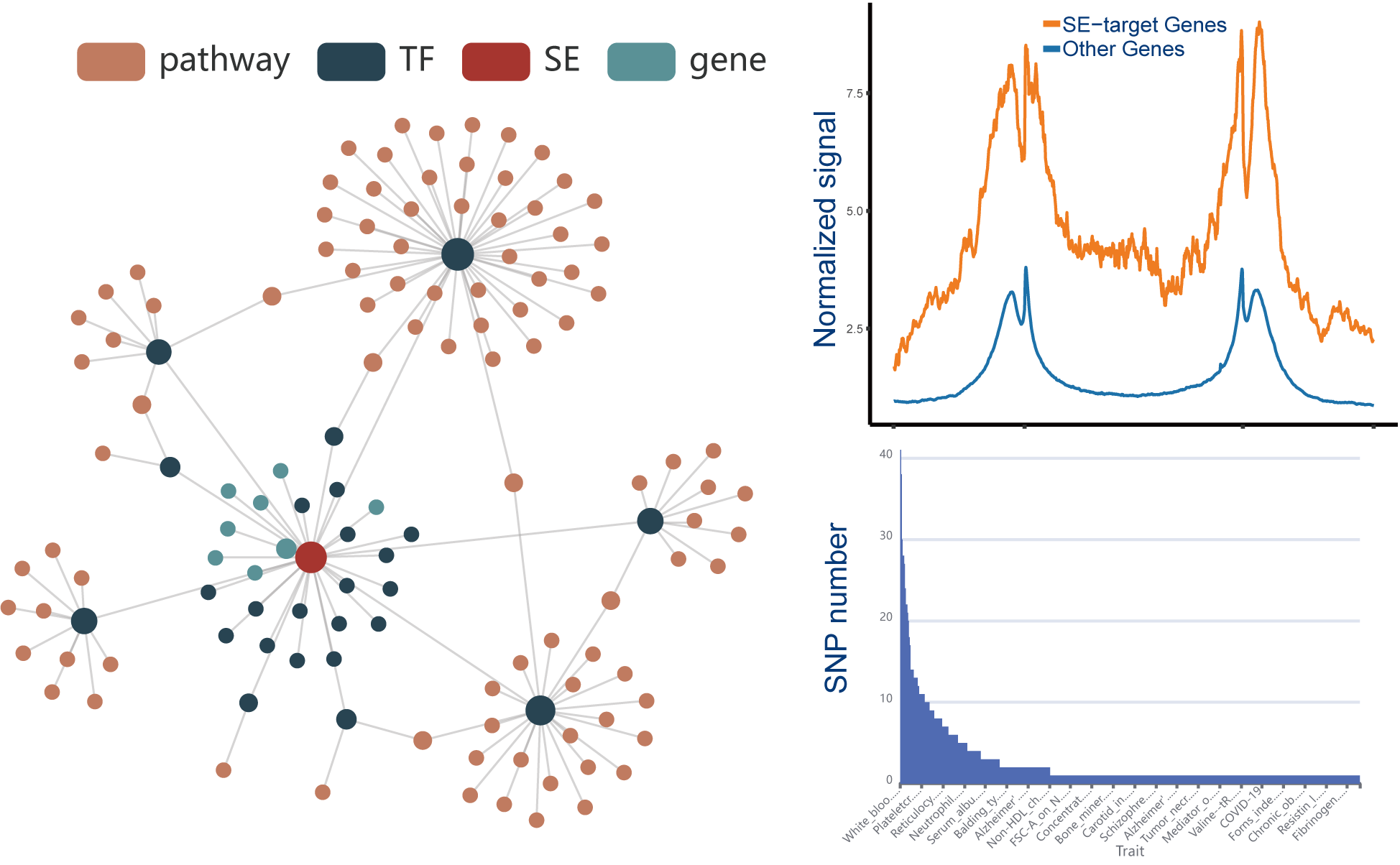
involving the identification of SE-associated genes, TFs occupying these SEs and the upstream signaling pathways of idenfitied TFs.
Users can perform the following SEs-associated analyses in the web server:
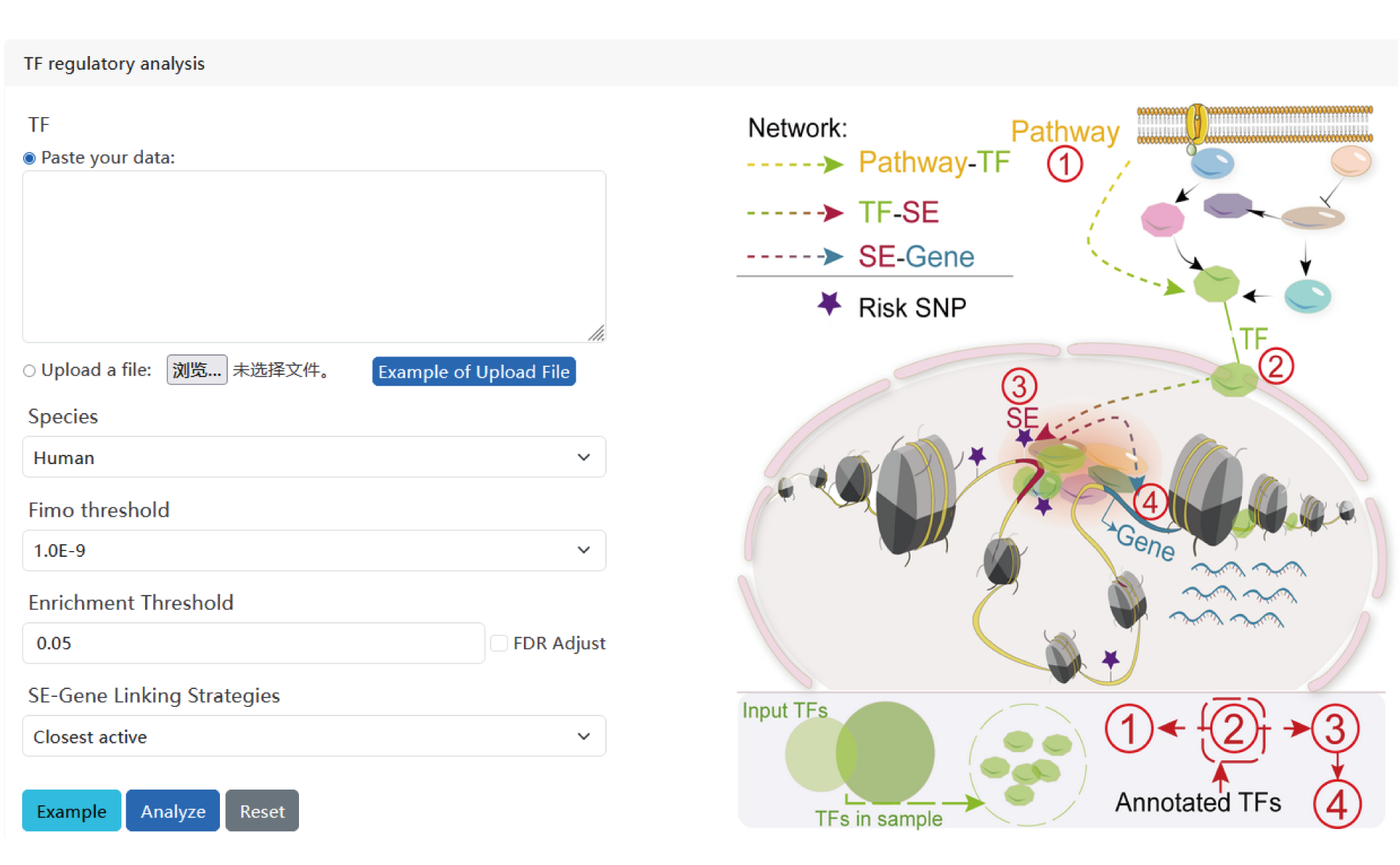
I. TF regulatory analysis.
II. Sample comparative analysis.
III. Pathway downstream analysis.
IV. Upstream regulatory analysis.
V. Genomic region annotation.
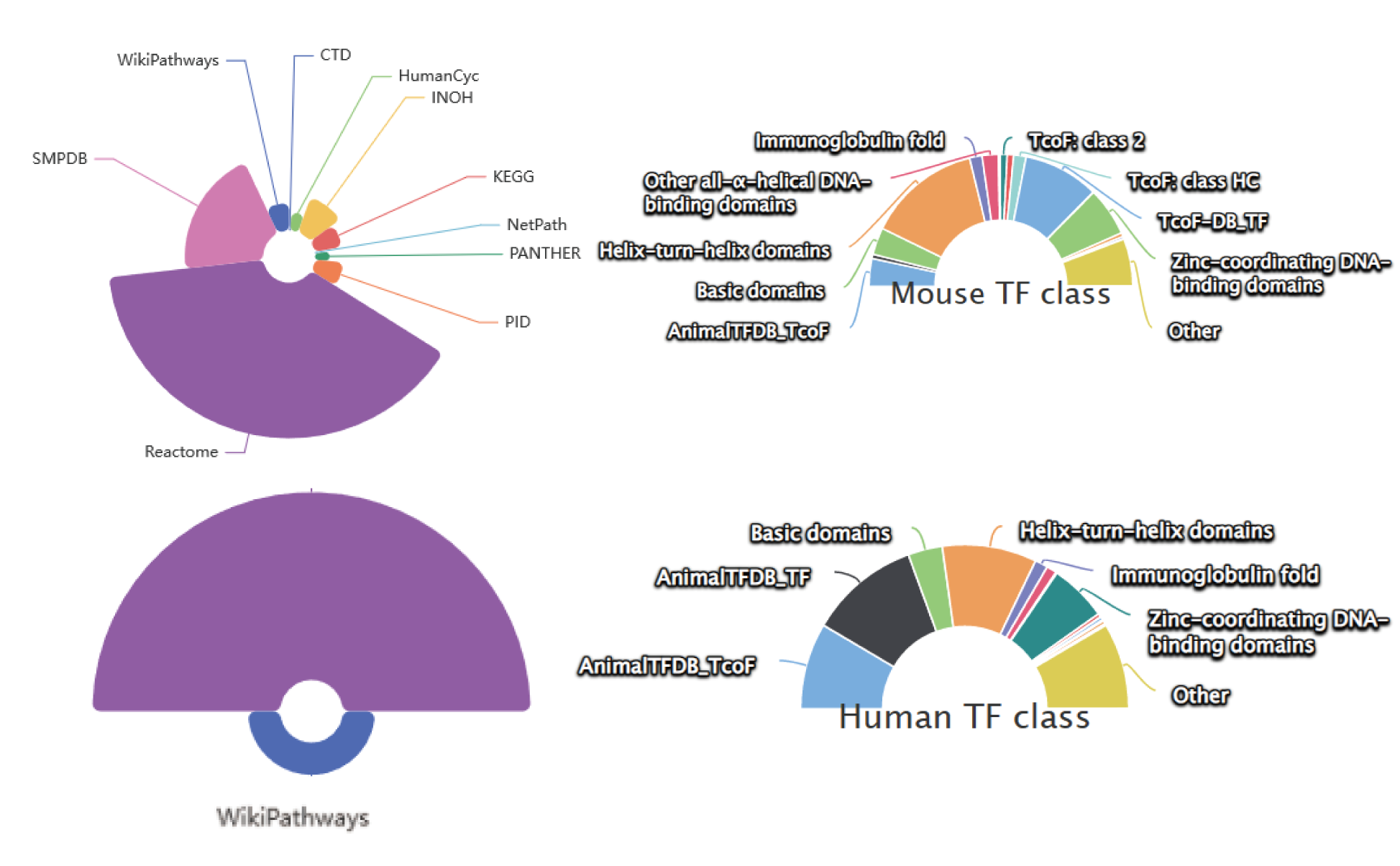
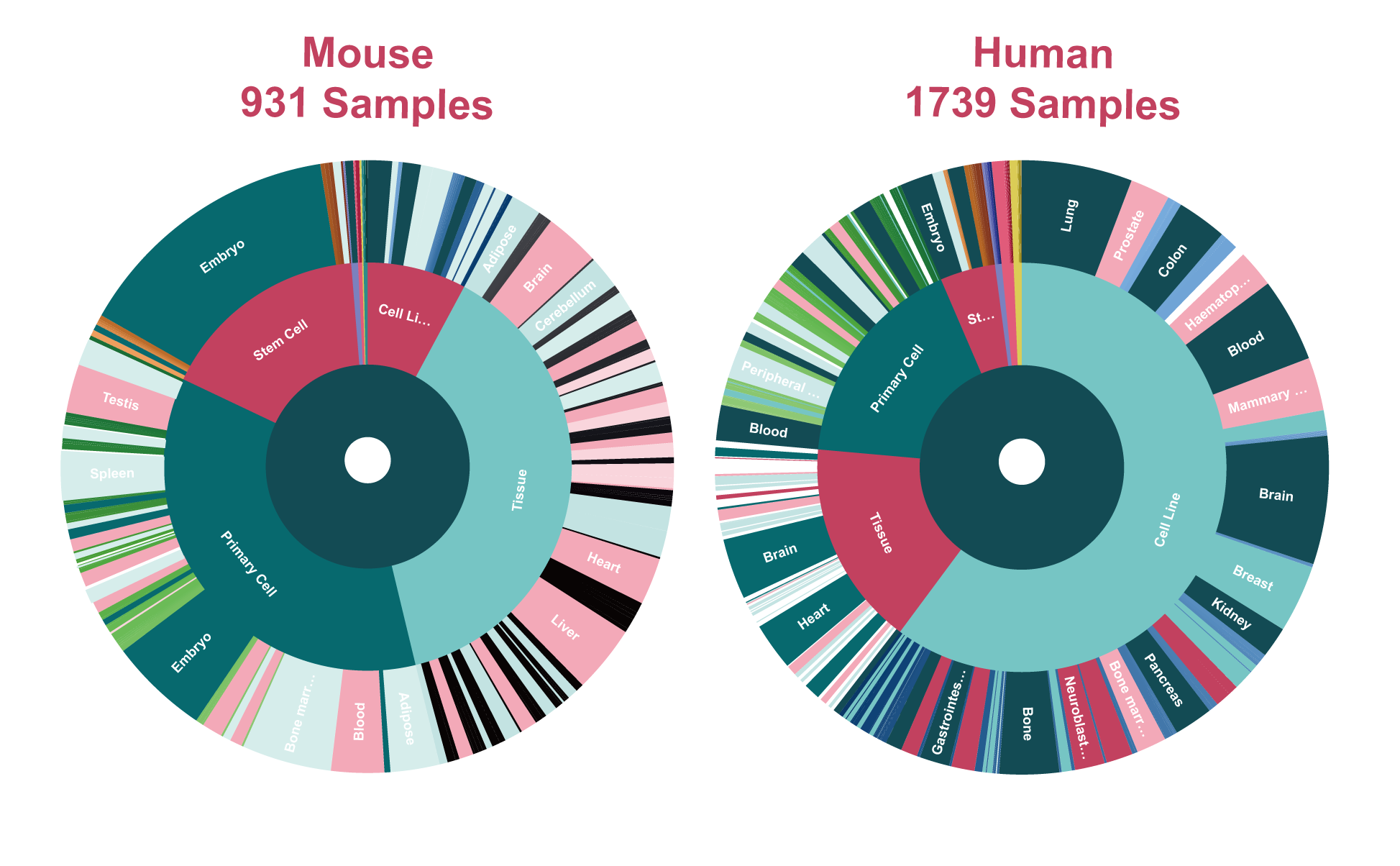
The current version of SEanalysis contains a total of 1,717,744 super-enhancers from over 2,670 Samples, including 1,739 human samples and 931 mouse samples; 10,710 TF ChIP-seq data for 1,468 human TFs and 446 mouse TFs generated from these cells/tissues; DNA-binding sequence motifs for 869 human TFs and 568 mouse TFs; as well as 2,880 human pathways and 1,487 mouse pathways. SEanalysis 2.0 provides risk SNP annotation to link super-enhancer with diseases/trait. SEanalysis 2.0 supports searching by either SEs, samples, TFs, signaling pathways or genes. Furthermore, the complex regulatory networks formed by these factors can be interactively visualized.
Statistics
Sample Statistics
| Human | Mouse | ||
|---|---|---|---|
| Pathway | The Number of Sources | 10 | 2 |
| The Number of Pathways | 2880 | 1487 | |
| TF(ChIP-seq) | The Number of Sources | 5 | 1 |
| The Number of TFs | 1468 | 440 | |
| The Number of Samples | 10 710 | 1051 | |
| TF(Motif) | The Number of Sources | 6 | 2 |
| The Number of TFs | 869 | 568 | |
| The Number of Motifs | 3680 | 742 | |
| SE | The Number of Sources | 5 | 5 |
| The Number of SEs | 1167518 | 550226 | |
| Samples | 1739 | 931 |
Live Statistics
For publication of results please cite the following article
Qian, F.C.*, Li, X.C.*, Guo, J.C.*, Zhao, J.M., Li, Y.Y., Tang, Z.D., Zhou, L.W., Zhang, J., Bai, X.F., Jiang, Y. et al. (2019) SEanalysis: a web tool for super-enhancer associated regulatory analysis. Nucleic acids research, 47, W248-W255.
Qian FC, Zhou LW, Li YY, Yu ZM, Li LD, Wang YZ, Xu MC, Wang QY, Li CQ. SEanalysis 2.0: a comprehensive super-enhancer regulatory network analysis tool for human and mouse. Nucleic Acids Res. 2023 Jul 5;51(W1):W520-W527.
SEanalysis 1.0 link is: http://www.liclab.net:8028/SEanalysis1/index.do
Sister Projects
|
The comprehensive human Super-Enhancer database |
|
|
A web tool for super-enhancer associated regulatory analysis |
|
|
An experimentally supported enhancer database for human and mouse |
|
|
A comprehensive human chromatin accessibility database |
|
|
A comprehensive database of human transcriptional regulation of lncRNAs |
|
|
A resource for transcriptional regulation information of circRNAs |
|
|
A comprehensive human gene expression profile database with knockdown/knockout of transcription factors |
|
|
A variation annotation database for human |
|
|
A comprehensive human lncRNA sets resource and enrichment analysis platform |