Cis-regulatory elements are important molecular switches in controlling gene expression and are considered as determinant hubs in the transcriptional regulatory network. Thus, it is urgent to collect and process cis-regulatory data to uncover the molecular mechanisms of cardiovascular diseases.
Here, we developed the Cis-Cardio database(http://8.218.144.187/Cis-Cardio/index.html), which aims to document a large number of available resources of cardiovascular-related cis-regulatory data, and to annotate and uncover the comprehensive mechanisms in cis-regulation level.
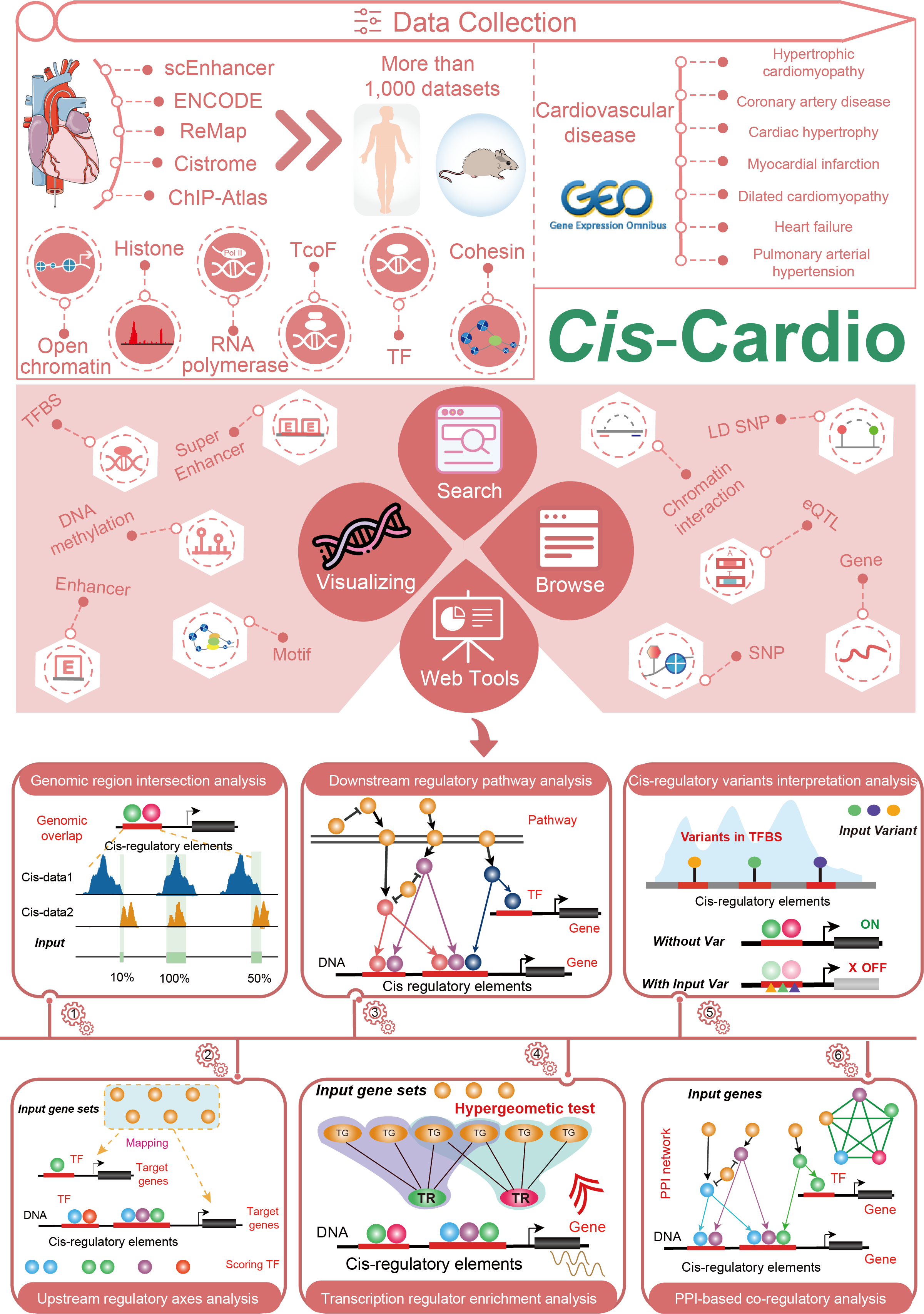
The current version of Cis-Cardio cataloged a total of 45,382,361 genomic regions from over 1,013 human and mouse epigenetic datasets, including scATAC-seq, ATAC-seq, DNase-seq, Histone ChIP-seq, TF/TcoF ChIP-seq, RNA polymerase ChIP-seq and Cohesin ChIP-seq. These datasets were manually curated from numerous of epigenetic databases and were almost covered all samples of cardiovascular systems, such as tissues, primary cells and IPS-induced cells.
Cis-Cardio provides the detailed and abundant (epi) genetic annotations in cis-regulatory regions, such as super enhancers, enhancers, TFBSs, methylation sites, common SNPs, risk SNPs, eQTLs, motifs, DHSs and 3D chromatin interactions. Cis-Cardio also provides cis-elements downstream target genes by mapping binding regions into genomes in two methods. Furthermore, Cis-Cardio provides various annotations for cis-elements target genes, including pathways, GO Terms and expression changes in major cardiovascular diseases. Especially, Cis-Cardio provides six types of cis-regulatory analyses for users, including Genomic region intersection analysis, Upstream regulatory axes analysis, Downstream regulatory pathway analysis, Transcription regulator enrichment analysis, Cis-regulatory variants interpretation analysis and PPI-based co-regulatory analysis.
4.1 Analysis
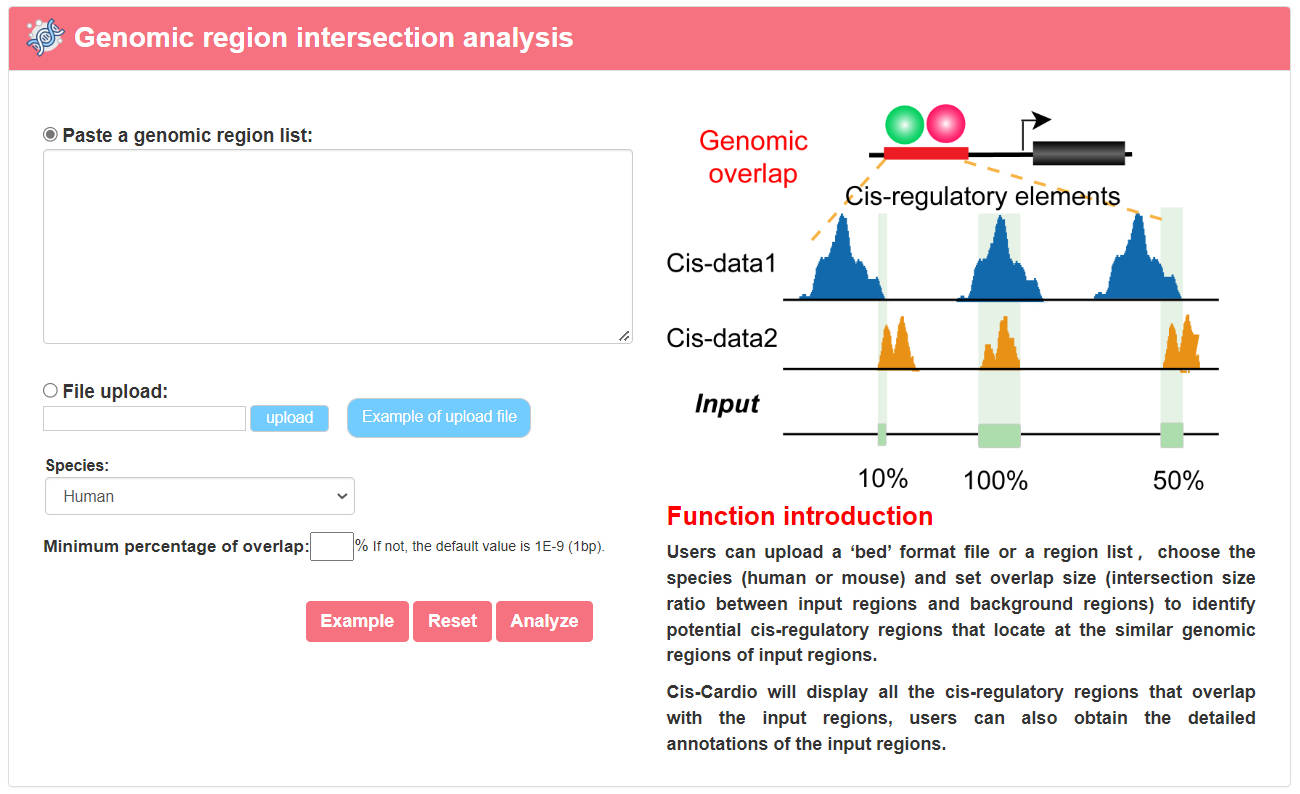
4.1.1 Genomic region intersection analysis
URL: http://www.licpathway.net/Cardio_cisDB/analysis/Analysis_peak.html
With the input of a genomic region list of interest or a ‘bed’ format file. Users can choose the species (human or mouse) and set overlap size (intersection size ratio between input regions and background regions) to identify potential cis-regulatory regions that locate at the similar genomic regions of input regions. Cis-Cardio will display all the cis-regulatory regions that overlap with the input regions, users can also obtain the detailed annotations of the input regions.
The guidance for input format is as below:
Region definitions are ordinary 3-field bed file format.
#column 1: The name of the chromosome (e.g. chr3, chrY).
#column 2: The starting position in the chromosome.
#column 3: The ending position in the chromosome.
For example: Example of Upload File
chr15:78540127-78540669
chr2:27628897-27629209
Brief information on the analysis results is displayed in a table on the result page, The peak can be visualized by clicking the "Peak ID" button. In the “Peak ID” page Cis-Cardio provides detial informations on the cis-regulatory regions, including peak overview, peak annotated information (enhancers, super-enhancers, risk SNPs, common SNPs, LD SNPs, eQTLs, DHSs, ATAC, 3D chromatin interactions, WGBS, TAD and CRISPR), target gene annotated (GO Term, pathway and disease) and expression changes of target genes in cardiovascular disease.

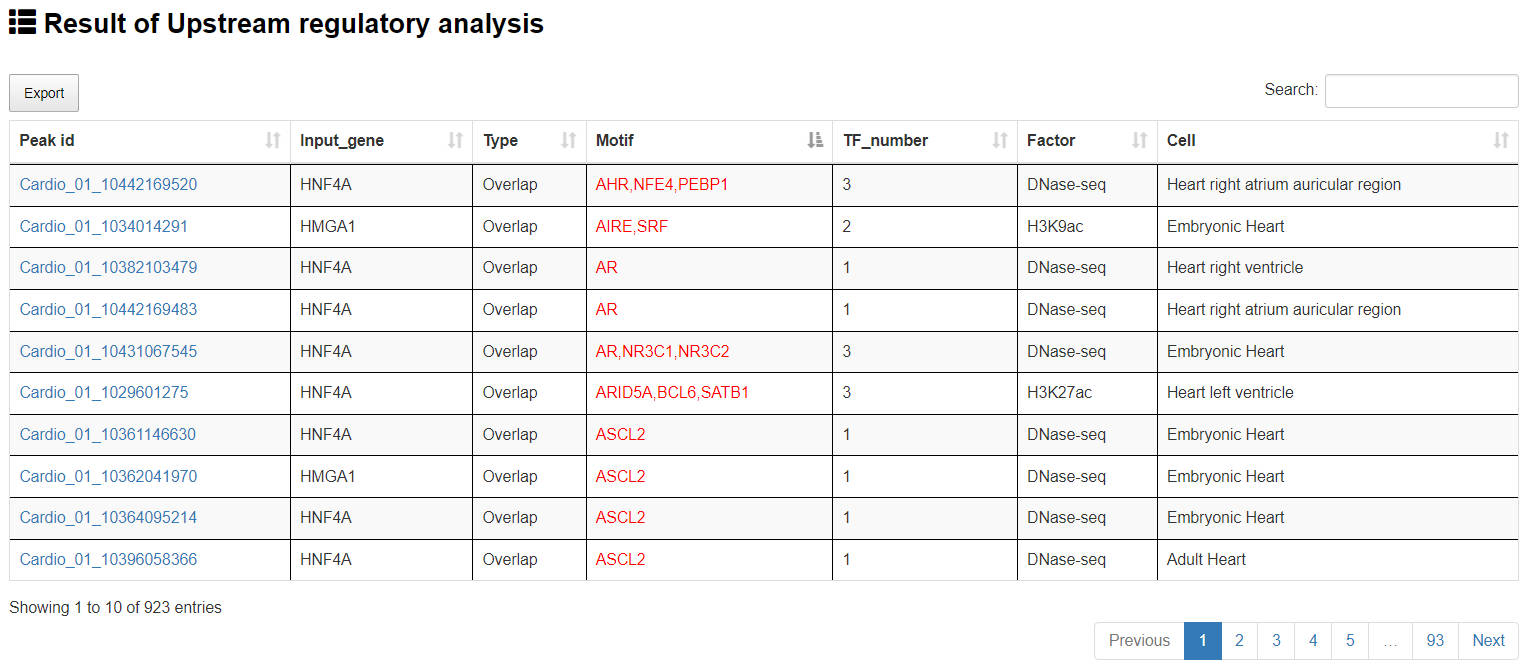
4.1.2 Upstream regulatory axes analysis
URL: http://8.218.144.187/Cis-Cardio/analysis/Analysis_gene.html
In this analysis, users can upload a ‘txt’ format file or a gene list, choose the species (human or mouse) and the cis-regulatory element-Gene linking strategies (ALL, GeneMapper_Overlap, GeneMapper_Closte, GeneMapper_Promixal and BETA). Cis-Cardio will map the target genes of all cis-regulatory elements, if the submitted genes are the target genes of a cis-regulatory element, Cis-Cardio will extract the upstream regulatory TFs of the cis-regulatory element and form the TFs/cis-regulatory elements/submitted genes regulatory axes.

The “Result of Upstream regulatory analysis” page presents cis-regulatory element overview and upstream regulatory TFs of cis-regulatory element. The cis-regulatory element can be visualized by clicking the "Peak ID" button various.
4.1.3 Downstream regulatory pathway analysis
URL: http://8.218.144.187/Cis-Cardio/analysis/Analysis_pathway.php
Users can submit a gene list, choose the species (human or mouse), select at least one pathway database, set p-value/FDR and set p-value of Fimo. Cis-Cardio will identify enriched pathways in up to the select pathway databases. In each pathway, Cis-Cardio will locate the terminal TFs and extract the terminal TF bound downstream cis-regulatory elements. Therefore, users can find the submitted genes/pathway/TFs/cis-regulatory elements regulatory axes.
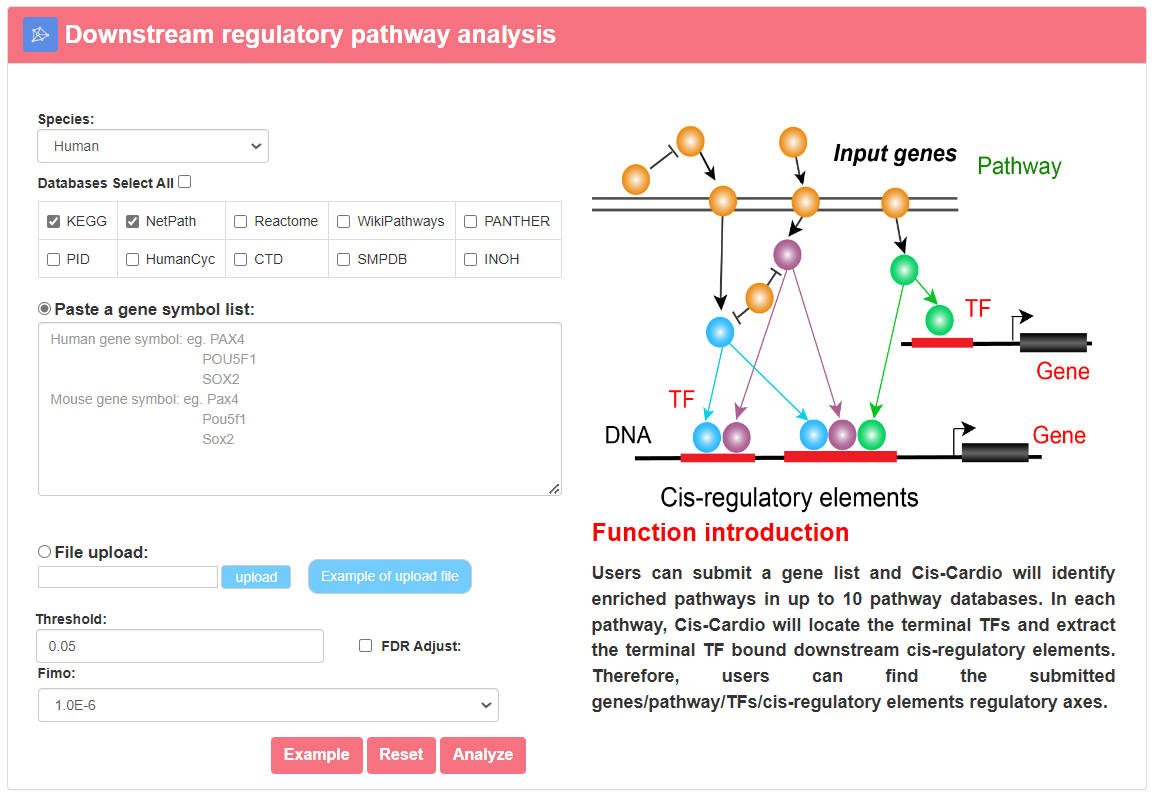
The output table shows pathway basic information (Pathway ID, Pathway name, Pathway source, Annotated gene, Annotated gene number, Total gene number, The terminal TF and TF number), enrichment score p-value and FDR. The pathways can be visualized by clicking the "pathway ID" button. The output table also provides a “Detail” page to further show the information of cis-regulatory elements. The regulatory networks based on the cis-regulatory elements samples in this pathway can also be visualized. The Peak can be visualized by clicking the "Peak id" button.

4.1.4 Transcription regulator enrichment analysis
URL: http://8.218.144.187/Cis-Cardio/analysis/analysis_enrichment/analysis_enrichment.php
In this analysis, users can input an interesting genome region list or a ‘bed’ file and choose the species (human or mouse). Cis-Cardio will calculate the significance of enrichment upstream transcription regulators by LOLA.

Cis-Cardio will return all significant transcription regulators (TR) information related the input regions to provide users with download and visualization. By selecting “UserSet”, Cis-Cardio provides the dataset overview, peak annotation and the peak annotation visualization.

Users also can submit a gene list to Cis-Cardio to identify the upstream transcription regulators (TR) based on hypergeometric test. The background TR-target gene pairs were constructed from each data.

Cis-Cardio will display all the datasets that overlap with the input genes, users can also obtain the detailed regulatory information of the enriched TRs.

4.1.5 Cis-regulatory variants interpretation analysis
URL: http://8.218.144.187/Cis-Cardio/analysis/analysis_snp/analysis_snp.php
Users can submit a variant name list (such as: rs10817286) to Cis-Cardio to extract the downstream peaks that locate the variant. Previous studies revealed that variants determine the transcription factor binding ability in DNA regulatory elements. Here, users can obtain the transcription factor binding sites (TFBS) in each cell-specific peaks via motif analysis.

The output table shows variant basic information (SNP ID and SNP Position) and peak information. The detail Motif information can be obtained by clicking the "Motif" button.

4.1.6 PPI-based co-regulatory analysis
URL: http://8.218.144.187/Cis-Cardio/analysis/analysis_ppi/analysis_ppi.php
It is important to decipher the regulatory axes of the regulatory proteins that locate in cell nucleus, which can exert functions by regulating transcription regulators. Here, users can submit a gene list (nuclear protein of interest) to find the direct transcription factor based on PPI network.

The output table shows the network topological importance score of the input genes and the downstream regulatory data of the interactive transcription regulators.

4.2 Browse
URL: http://8.218.144.187/Cis-Cardio/analysis/Analysis_peak.html
The ‘Data-Browse’ page is organized as an interactive and alphanumerically sortable table that allows users to quickly browse samples and customize filters through ‘Species’, ‘Factor type’ and ‘Cell type’. Users may further click on the ‘ID’ to view cis-regulatory elements for a given sample.

4.3 Search
URL: http://www.licpathway.net/Cardio_cisDB/search.php
Cis-Cardio provides four query search methods for users to obtain the cis-regulatory data, including ‘Search by Factor’ (select species and input factor name of interest), ‘Search by gene’ (select species and input gene symbols of interest), ‘Search by genomic region’ (select species and input genomic position) and ‘Search by tissue/cell’ (select species and input tissue or cell name of interest).

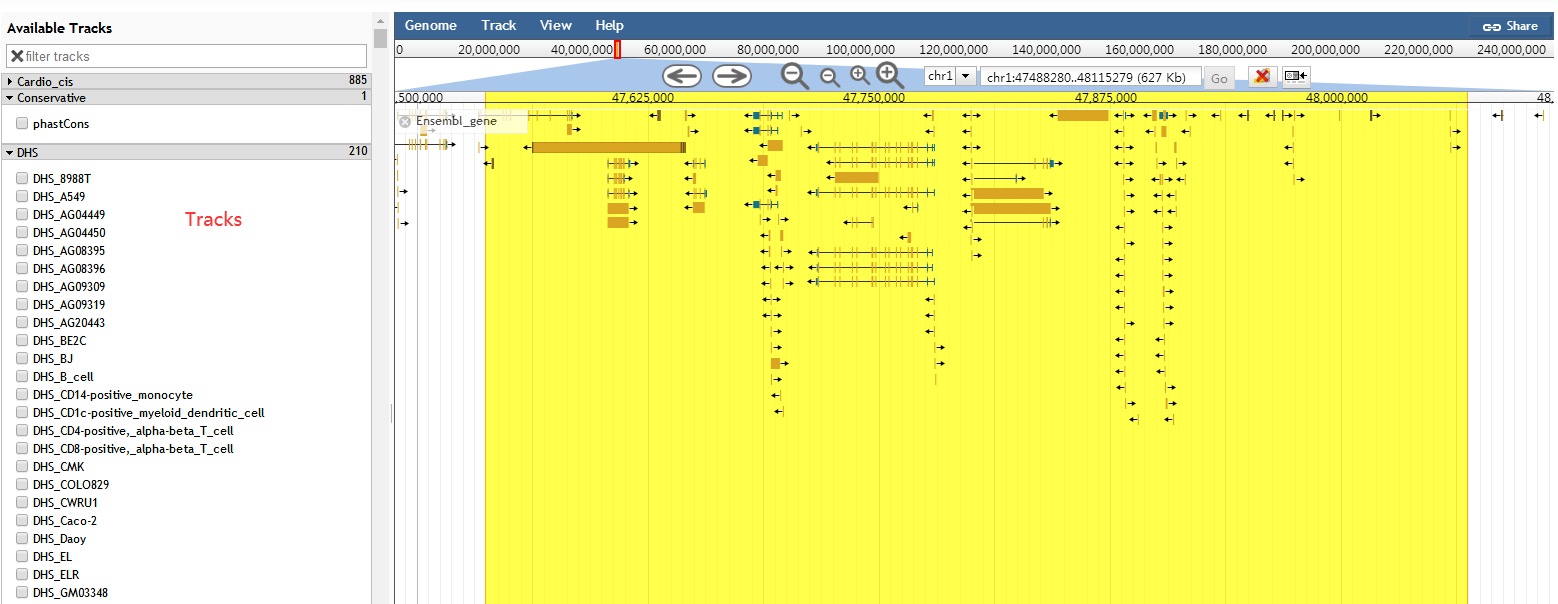
URL: http://www.licpathway.net/Cardio_cisDB/Genome_Browser.php
In order to help users to intuitively view proximity information of cis-regulatory elements in the genome, we developed a personalized genome browser using JBrowse and added a lot of useful tracks such as cis-regulatory regions, enhancers, super-enhancers, TFBSs, DHSs and genes. In addition, Cis-Cardio can exhibit pie charts of chromosome distribution and the histograms of the number statistics of annotation information in cis-regulatory regions.
URL: http://8.218.144.187/Cis-Cardio/download.php
Cis-regulatory data information, cis-regulatory elements of all samples and downstream target genes are provided for download in the ‘Download’ page. Users can quickly download information of interest. We support ".bed" or ".txt" formats.

The current version of Cis-Cardio was developed using MySQL 5.7.17 (http://www.mysql.com) and runs on a Linux-based Apache Web server (http://www.apache.org). PHP 7.0 (http://www.php.net) was used for server-side scripting. The interactive interface was designed and built using Bootstrap v3.3.7 (https://v3.bootcss.com) and JQuery v2.1.1 (http://jquery. com). ECharts (https://www.echartsjs.com/) and Highcharts (https://www.highcharts.com.cn/) were used as a graphical visualization framework. We recommend to use a modern web browser that supports the HTML5 standard, such as Firefox, Google Chrome, Safari, Opera or IE 9.0+ for the best display.
The Cis-Cardio database is freely available to the research community using the web link (http://8.218.144.187/Cis-Cardio/index.html). Users are not required to register or login to access features in the database.